
- Core Center:Microbe Division/Japan Collection of Microorganisms (JCM), RIKEN BioResource Research Center
- Principal Investigator:Moriya Ohkuma
- FAX:+81-29-836-9561
概要Overview
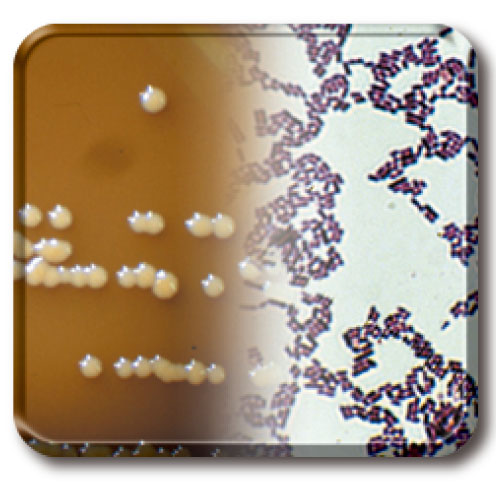
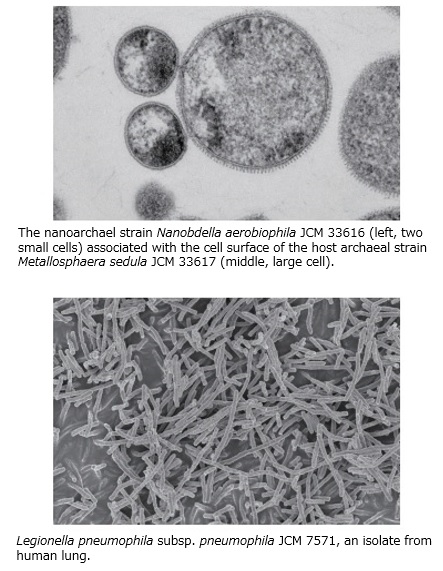 The Japan Collection of Microorganisms (JCM) provides diverse microbial strains of bacteria, archaea, yeast, and filamentous fungi. We take much care for quality control and will continue to ensure the high credibility of our resources and services. We also provide highly valuable microbial strains that have been reported in scientific publications. JCM strains are widely used and contribute to a variety of research fields related to environmental and health science as well as general microbiology.
The Japan Collection of Microorganisms (JCM) provides diverse microbial strains of bacteria, archaea, yeast, and filamentous fungi. We take much care for quality control and will continue to ensure the high credibility of our resources and services. We also provide highly valuable microbial strains that have been reported in scientific publications. JCM strains are widely used and contribute to a variety of research fields related to environmental and health science as well as general microbiology.
Stock
・Bacteria: about 21,000 strains
・Archaea: about 1,000 strains
・Fungi: about 8,800 strains
・Type or type-derived strains: about 10,000 strains
Subjects in the NBRP programs related to “General Microbes”
【 Value addition subprogram/ Genome Information Upgrading Program 】
| FY2023-FY2024 | 難培養性原核生物の基準株の完全長ゲノム情報整備 (in Japanese) |
| FY2014 | Genome sequencing of eukaryotic microorganisms of NBRP general microbes |
| FY2012 | Genome sequencing of microbial strains for environmental and health science |
【 Technology development subprogram/ Fundamental Technology Upgrading Program 】
| FY2020-FY2021 | Development of reference MALDI-TOF MS data that enables rapid identification of various microbes(in Japanese) |

